Research Projects
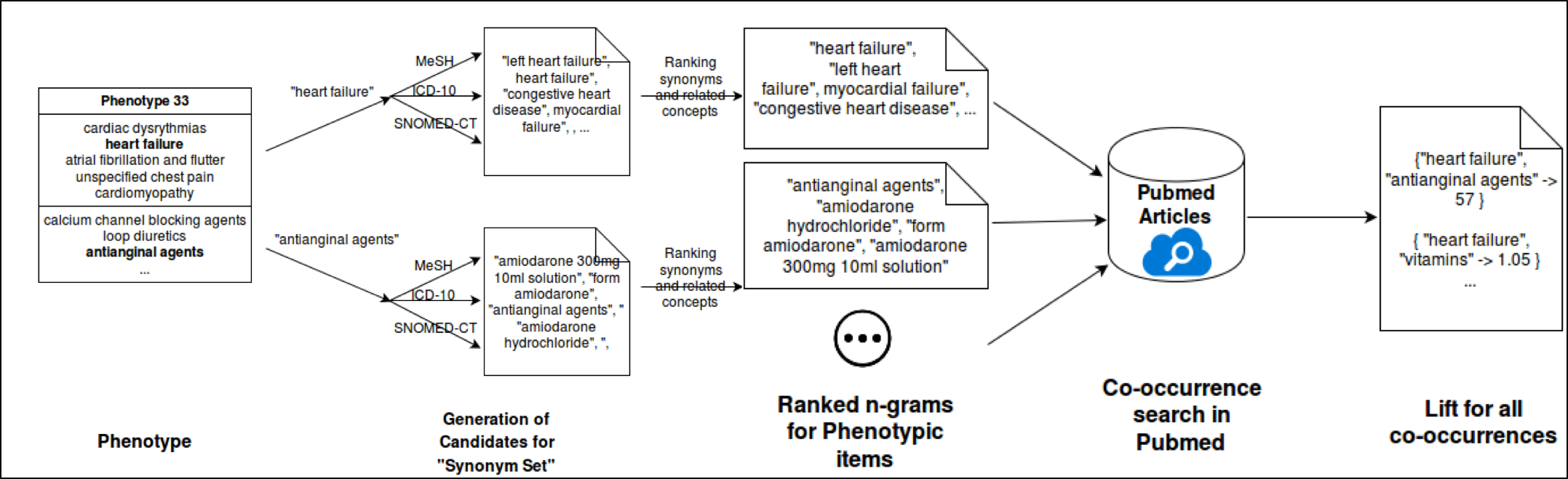
Pheknow-Cloud
Our work focuses on developing a tool to help evaluate high-throughput phenotype candidates using PubMed, an online repository of medical literature. We use co-occurrence analysis to build sets of evidence for user-supplied candidate phenotypes
Collaboration with UT-Austin and Northeastern.
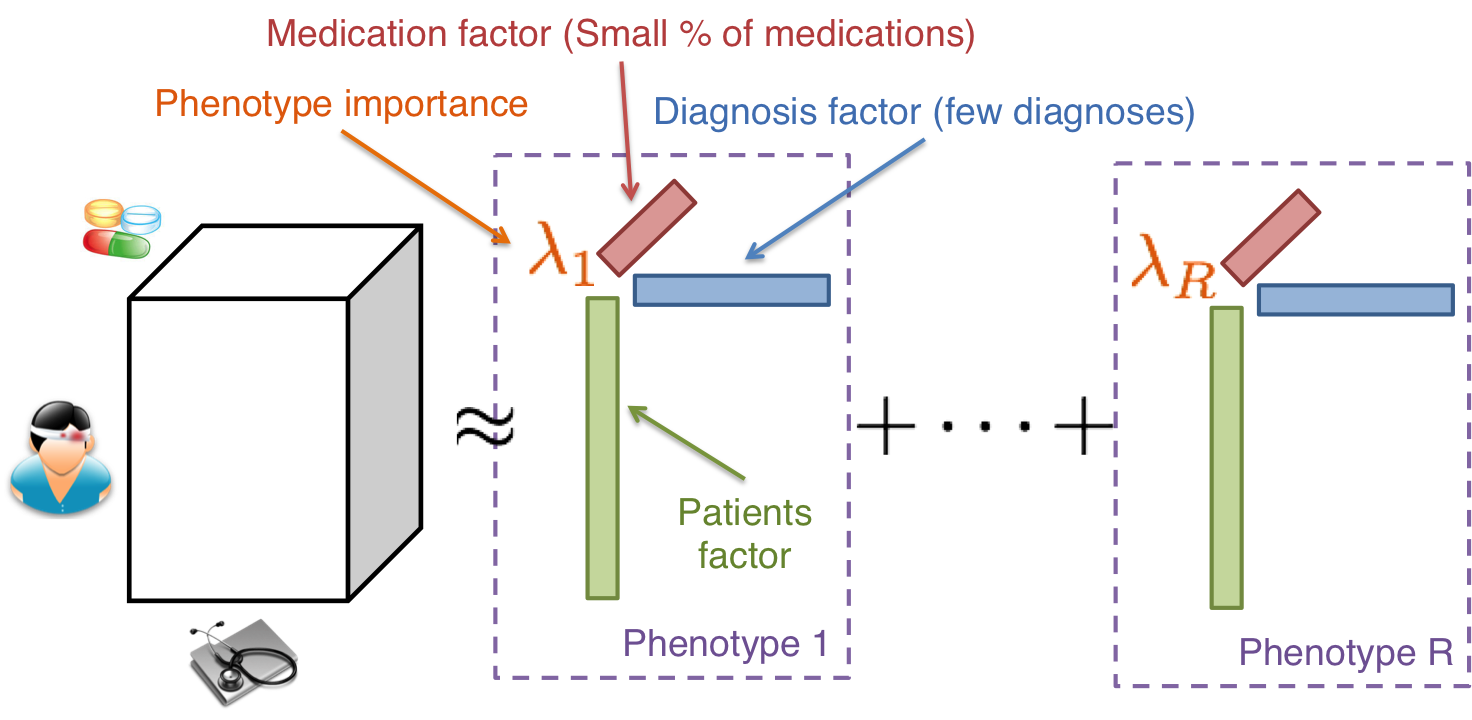
High-throughput Phenotyping on Electronic Health Records using Multi-Tensor Factorization
Our research addresses the problem of transforming raw electronic health record (EHR) data to medical concepts with minimal human intervention. We posit the use of multi-relational tensor factorization approaches to generate concise and clinically relevant phenotypes. The proposed computational framework will provide powerful, data-driven, and interpretable approaches for transforming high-dimensional EHR data into medical concepts.
Collaboration with UT-Austin, Georgia Institute of Technology, Vanderbilt University, and Northwestern University.

Medication usage amongst CFS patients
Chronic Fatigue Syndrome (CFS) is a complex and devastating illness of unknown cause and origin. Patients are often on multiple medications to manage the symptoms. We use tensor factorization to improve our understanding of CFS as they provide a natural framework for flexible representation and analysis of multi-aspect data.
This is an ongoing collaboration (started as a summer internship) with Jin-Mann (Sally) Lin and Brian M. Gurbaxani from the CDC.

High-throughput phenotyping via tensor factorization
EHR based phenotyping is the process of mapping EHR data to meaningful medical concepts, where each concept contains a set of clinical features. We developed a nonnegative tensor factorization method to derive phenotype definitions with virtually no human supervision, representing the data source interactions naturally using tensors (a generalization of matrices). The resulting tensor factors automatically reveal patient clusters on specific diagnoses and medications.
This work was performed during my summer internship in the Healthcare Analytics group, mentored by Jimeng Sun.
DYNACARE: Dynamic cardiac arrest risk estimation
A semi-supervised time-series framework to predict the cardiac arrest time for high-risk ICU patients. A single latent process is used to represent each patient's cardiac arrest risk and the cardiac arrest event is defined as a temporal signature in the latent process. Our algorithm is inspired by financial econometric and yields interpretability and predictability of a cardiac arrest event.
This work is a collaboration with Yubin Park and Carlos Carvalho.

Multiple Sclerosis Risk Prediction Model
Multiple sclerosis (MS) is a chronic, disabling autoimmune disease that affects the central nervous system. Early diagnosis and treatment of the disease have been shown to be effective at slowing the development of disabilities. However, early MS diagnosis is difficult because symptoms are intermittent and shared with other diseases. We investigated the use of commonly available data in electronic medical records to create a risk prediction model; thereby helping clinicians perform the difficult task of diagnosing an MS patient.
This work was performed during my summer internship in NorthShore Clinical Research Center, mentored by KP Unnikrishnan.
Septic shock prediction for patients with partial observations
Sepsis is one of the leading causes of mortality in intensive care unit (ICU) patients. However, development of highly accurate predictive models for medical applications is often complicated by the nature of clinical data, which are typically noisy and inconsistently gathered. Our work investigates the role and impact of imputation methods while building predictive models for septic shock. By limiting our features to commonly observed, mostly non-invasive clinical measurements, we build better predictive models that are generalizable to broader groups of ICU patients.
This work was performed in collaboration with Cheng H. Lee.

PhysioNet 2012 Challenge
The focus was to develop methods for patient-specific prediction of in-hospital mortality. We used 42 variables (37 time-varying measurements) collected during the first two days of an ICU stay to predict which patients survive their hospitalizations, and which patients do not. Our entry placed in the top 10 (of 23) for both evaluation categories.
Our team included Cheng H. Lee and Natalia M Arzeno.